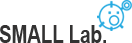Nanoparticle Chemistry
To establish a platform technologies enabling simultaneous imaging, tracking and extracting bio-physical parameters in live cells, We apply the power (optical stabilities) and of inorganic-organic nanomaterials.
Recently, we were able to control the spatial distribution of Notch receptor with single/sub- cellular resolution, and activate Notch signaling spatially, temporally, and quantitatively via magnetic mNPs (Seo et al., Cell, 2016). For other proteins, such as growth factor receptors and ion channels, however, we need develop appropriate tools to turn on the each signaling by establish multiple perturbation system including molecules induce the conformational change of the proteins, nanoparticles exert chemical, mechanical, thermal energy to proteins.
We are also expanding nanoparticle technology to utilize the optical and catalytic properties of hybrid nanoparticles to monitor catalytic reaction in real time. For example, we are attempting to develop highly sensitive catalytic reaction monitoring system, designed the structure composed of plasmonic materials with metal oxide as a model system for photochemical reaction.
Single-Molecule Biophysics
To understand how cells exploit different stimuli and signaling pathways are combined and orchestrate to control a diverse array of cellular processes in widely different spatial and temporal domains, we are developing imaging and manipulating nanotechnology for receptor dynamics and their functions (e.g. EGFR, NMDAR, PV-1, and PD-L1) in spatiotemporal and quantitative way.
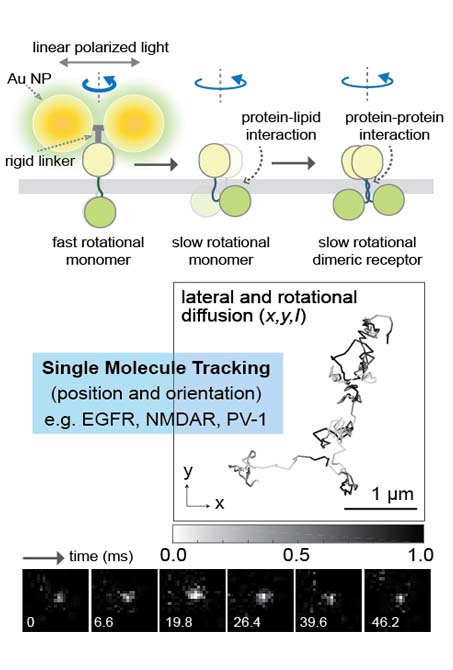
Living cells use genetically encoded molecular networks to monitor their environment and make sophisticated decisions. Cellular networks, or various signaling pathways are interconnected and multifunctional, we are developing imaging and tracking method of cell signaling (e.g. Erk, Akt)in live cell on appropriate spatiotemporal scales.
Monitoring the dynamics of both of receptor protein and cell signaling in live cell on appropriate spatiotemporal scales may provide key information on long-standing questions in molecular and cellular regulatory mechanisms. However, tools capable of imaging the dynamic changes over time have been elusive. We, therefore are developing microscopy (i.e. super-resolution fluorescence and dark-field) and spectroscopy (i.e. single molecule spectroscopy) technology.
An artificial-intelligence method provides powerful tools for analysing and interpretating our observed big size of data. We are using simple Machine learning, deep-learning algorithms to understand not only diffusive- conformational dynamics of proteons and cell signaling, but also catalytic reactivities in chemical reactions.
© 2016 SMALL Lab. Made with by D.Seo